I’ve been working on quite a few things recently, and will get back to regular posts soon. In the meantime, here’s a brief introduction to Group Analysis tests in AFNI.
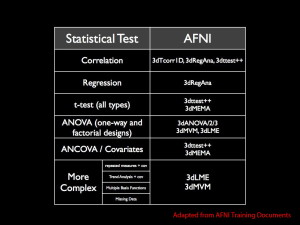
I see the information related to Group Analyses falling into two categories: 1) Which program to run and when; 2) all the other stuff related to Group Analyses beyond just running one program to compare X. Today I’m going to outline the programs one would use for doing different types of Group (sometimes also called 2nd-level) analyses. To aid this, I’ve adapted an AFNI table and attached it below.
As you can see, for just about any type of group level test, there exists one (and sometimes MANY) different AFNI programs. The choice of which program to use can depend on what you’re already comfortable with or what you really need for a particular analysis. For example, you can use 3dRegAna to do regression, or ANCOVA. But an easier way to do an ANCOVA is to use 3dttest++ or 3dMEMA because in 3dRegAna you must specify each variable in the command; whereas in 3dttest++/3dMEMA you can use a text file to list out your covariates and have them all run simultaneously to view the impact of each covariate on your model.
Another reason to choose one program over another is speed. You can perform ANOVAs (one-way or multi-factorial) in 3dANOVA (and it’s cousins 3dANOVA2 and 3dANOVA3), or you can do this same analysis in 3dMVM (MVM = Multivariate Modeling). The advantage of 3dANOVA/2/3 is speed, these programs run incredibly fast! The disadvantage of 3dANOVA/2/3 is that they require balanced designs (no unequal groups) and they cannot take covariate information. In swoops 3dMVM, which can handle both unbalanced designs and covariates, but is considerably slower.
I will also take a moment and recommend that everyone look through the help information on gen_group_command.py, which can automatically generate the necessary commands for 3dttest++, 3dMEMA, 3dANOVA, 3dANOVA2, and 3dANOVA3 based on the input files (passed to the script) and some additional information, which can be a huge time saver.
gen_group_command.py -command 3dttest++ \
-write_script ttest_example.tcsh \
-prefix condition_${name}.nii.gz \
-dsets subject??.nii.gz \
-subs_betas 'conditionA#0_Coef'
Just that will generate a script to run 3dttest++ (saved to ttest_example.tcsh). That script will run a single-sample t-test on the data files in the folder that are labeled subjectXX.nii.gz, where XX are two numbers, for a particular condition. In addition to being a huge time saver for generating one script, this is also very helpful for allowing you to quickly add or remove subjects to the t-test (please use statistics responsibly!).