So far in the DTI series we’ve covered how to process data entirely in AFNI (Part 1, Part 2). We’ve also covered how to preprocess and fit tensors in the nicely packaged form of TORTOISE (Part 1, Part 2, Part 3). Regardless of how you got to the point of having tensors fit, you can continue to process data in AFNI! And one of the ways that you can use AFNI to analyze your data is to generate tractography results using the tool 3dTrackID. But before you go running deterministic or probabilistic tractography, I want to take a moment and say that it’s still important to double check your data! Yes, I know that I told you that TORTOISE does all kinds of wonderful things, and many people reading this blog may think that AFNI can do no wrong. But even if those two things were true, your data can still be lying to the software!
Which brings me to the topic of today’s post. Flipping. No, I’m not talking about gymnastics or hand gestures. Instead I’m talking about how your B-MATRIX (sometimes bvec file) is setup in relation to your data. It’s entirely possible that despite what you may believe, the DICOM or NIFTI files you are using might be lying to you. By lying, I mean that the B-MATRIX/bvecs/whatever doesn’t match what’s really going on with your data. So before you start running tractography assuming your data is correct, take a few minutes and double check your data using these steps.
First of all, take whatever data you have and run a whole-brain deterministic tractography search on it using 3dTrackID (here my naming conventions match TORTOISE):
3dTrackID -mode DET \ -logic OR \ -mask MASK.nii \ -netrois MASK.nii \ -dti_in INPREF_ \ -prefix o.dti.det.test
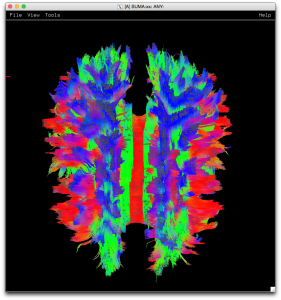
This will generate a whole-brain DTI image (shown below) that you can view in SUMA using:
suma -tract o.dti.det.test_001.niml.tract
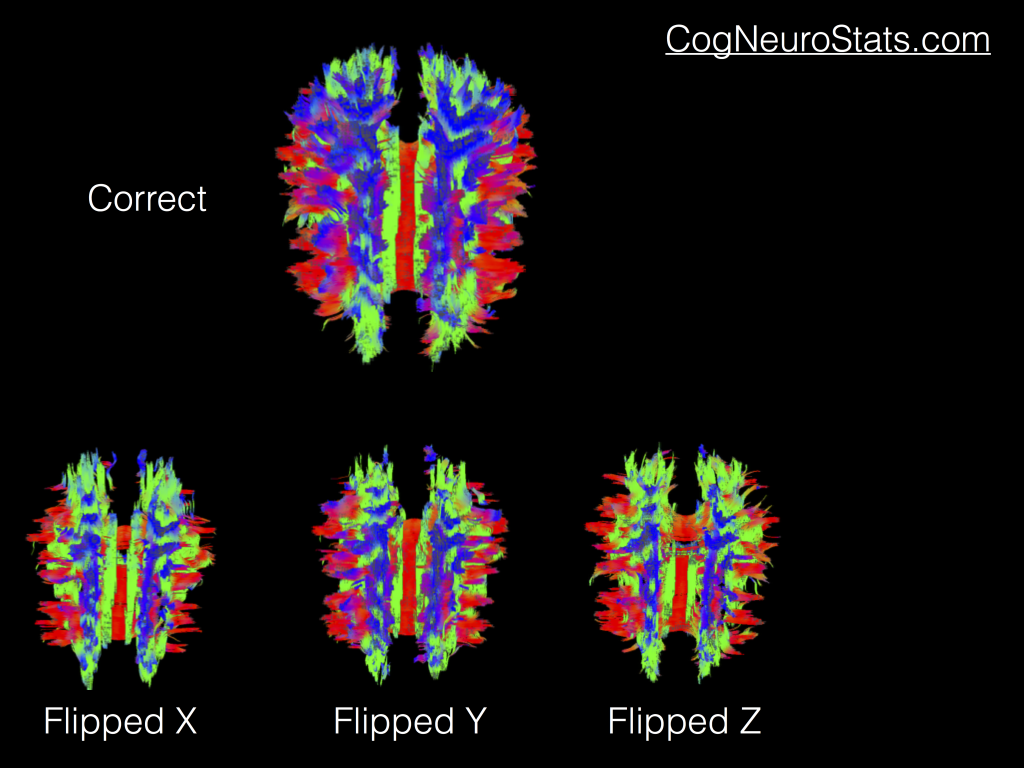
As you can see, we have a pretty healthy looking DTI image. But if your data didn’t match the B-MATRIX that you were dealing with you might see some of these common changes (more available on SUMA’s new fancy documentation).
Admittedly the hardest one to differentiate from the correct results is the “Flipped Y”. But truthfully you may end up playing with all of these flips (or even combinations of flips) until you find one that works for your data. So what does that mean? Well it means you’re going to get to dust off a couple programs that you might not use on a regular basis: 1dDW_Grad_o_Mat and 3dDWItoDT.
To make things even more concrete, let’s take an example where the data is flipped in the X direction. If I’m using the output of TORTOISE, the process I would use is as follows. My files are listed as DWI.nii, BM.txt, and possibly a MASK.nii. These are the diffusion weighted data, a whole-brain mask (if you don’t have one, use 3dAutomask) and the B-MATRIX (if you used the AFNI export in TORTOISE it’s in the AFNI format referred to in 1dDW_Grad_o_Mat).
#Flip the gradient directions in your BM.txt 1dDW_Grad_o_Mat \ -in_bmatA_cols BM.txt \ -out_bmatA_cols BM_flipX.txt \ -flip_X \ -keep_b0s
#Refit your tensors with the new gradients
3dDWItoDT -prefix DT_flipx \
-reweight -nonlinear -eigs \
-sep_dsets \
-mask MASK.nii \
-bmatrix_Z BM_flipX.txt DWI.nii
#Use whole-brain deterministic tractography to check
3dTrackID -mode DET \
-logic OR \
-mask MASK.nii \
-netrois MASK.nii \
-dti_in DT_flipx_ \
-prefix o.afni.flipx
#visualize in SUMA
suma -tract o.afni.flipx_001.niml.tract
And if that didn’t work, try flipping another direction. Here is another place where scripting is your friend. Or enemy if you made a typo, but we’ll hope that it saves you time! See you next time!