AFNI has three major avenues to running a single subject analysis. 1) You can use uber_subject.py to configure the analysis in a Graphical User Interface (GUI); 2) You can use afni_proc.py to specify the analysis using the command line; 3) You can write your own command line commands in your own script, which calls each AFNI program as needed with the settings that you desire.
The major problem with approach #3 is that AFNI has thousands of options across more than 100 programs! This is part of the reason why AFNI had such a steep learning curve. But fortunately, options #1 and #2 are considerably easier for both the new AFNI user and the experienced AFNI user alike. In quick summary, uber_subject.py provides a GUI interface for setting up your analyses. Underneath that GUI, uber_subject.py calls another python script called afni_proc.py. Now here’s an important distinction – afni_proc.py has many many many more options than uber_subject.py for configuring your analyses. But try as I might, for easily 75% of my analyses, uber_subject.py has all of the options you will need turned on by default.
And with that, let’s look at uber_subject.py! The program itself comes installed with AFNI, but to get everything working requires the installation of PyQT. If you want some guidance on that, checkout the previous post for Mac Installation instructions. Typing “uber_subject.py -help_install” on any platform will give you the AFNI group’s recommendations for installing the necessary dependencies. Launch uber_subject.py by typing its name into the terminal window on a Mac or Linux and be presented with the following:
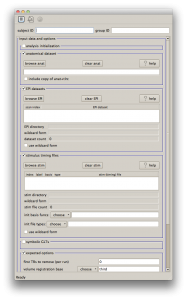
Notice the scrollbar on the right side indicating more options below! In addition, clicking on checkboxes to add options may increase the length of the scrollbar. Let’s fill in the options with some reasonable defaults (some people may find other defaults more reasonable than these). These options will setup AFNI to perform outlier detection, slice timing correction, perform a single transformation aligning the EPI to the high resolution anatomical with motion correction and the warp to MNI space, spatial smoothing, masking, converting the scanner units into standard units (for % signal change), and running the regression via 3dDeconvolve with motion parameters entered in.
Subject ID: A subject ID or Name or identifier
Group ID: If you have multiple groups, use the name here, otherwise “all” or the name of the study works well
Analysis Initialization: This allows you to set analyses for either “task” or “resting” and let’s your specify analyses in the volume or on the surface. For most analyses, I tend to start with a “task” and in the volume. In part because the surface requires a good FreeSurfer or Caret cortical surface. If you are new to AFNI, leave the processing blocks to the default.
Anatomical Dataset: This is your high resolution image (think MRPAGE or SPGR).
EPI datasets: These are your function EPIs.
Stimulus Timing Files: these are AFNI specific timing files. They should be in text format with one file per stimulus condition and one line in each file per run. So if you have two runs, you would have two lines in each stimulus file. If you have a stimulus condition that only appears in one run but not the other, add a * to let AFNI know that it has a blank line. After you add them to uber_subject.py, add a label by clicking on the empty label box and typing one in.
Symbolic GLTs: Are your “contrasts”, here you can specify the difference between Condition A and Condition B or any other combination that you wish. If this seems daunting, click the “init with examples” to see some generic contrasts that relate to your stimulus labels.
Expected Options: These are a series of useful options. First TRs to remove include your pre-steady state images, but only if your stimulus timing files reflect the removal of these. Volume register base options are first, third, and last. Since I trim off the first several TRs on my imags, I tend to use first. Blur size is important, start with two voxels (in my case this is 4mm) if you’re new and then google blur size impact on fMRI. If you’re an old hat, fill in your value here. Motion censor, I would stick with the defaults.
Extra Regress Options: Change the outlier censor to 0.1 (censors a TR if 10% of voxels are outliers). Jobs can be set to the number of processors on your computer. Even if you have 24 cores, I wouldn’t really set this higher than 12. GOFORIT should stick with the defaults. bandpass is usually left blank. I also add the 3dREMLfit option.
Extra Align Options: I use the default LPC, but add “use giant_move” as the high resolution image is typically in a very difference space than the EPI.
Extra tlrc options: These are additional options for the warp to standard space. I use the defaults of TT_N27+tlrc and the option to strip the skull. You can view the different templates and make your own decision there.
Now that you’ve specified all of the options, click the button in the upper left that looks like a text document in a rectangle. This will show two text windows, the first shows you the script that will be run, notice that it calls afni_proc.py. The second shows the variables that uber_subject.py used to construct the script. Importantly, you can modify this generated script if you want to add or change options. Just remember to save with command or control + S. To run the script press the green circle button in the upper left of the uber_subject.py window. A progress window will show up to show you your analyses being run.
Now that you’ve run one subject, repeat the process for all of your other subjects. Alternatively, copy the afni_proc.py script generated by uber_subject.py and modify it for each subject. After you’ve run all of your subjects, you can do group analyses with a number of tools to be described later – including uber_ttest.py.