There just isn’t enough time in the day to get everything done. And the more time that I spend on getting little things done, the less time I have for the “big picture”. Big picture will include writing papers, chapters, and grants. The ability to do repetitive tasks quickly and automatically is probably the most important skill to develop early on in your career. One way to accomplish this is to hire minions. The more sensible way is to learn how to program.
There are a lot of programming languages in the world. Some are very low-level (e.g. C, C++), requiring you to specify quite a few details. Other languages are very high level (e.g. Python, Ruby), which take care of a lot of the grit for you, while sacrificing some of the flexibility of lower-level languages. But one of the most overlooked and easy to develop skill for someone dealing with imaging data is the ability to script in the shell of your UNIX/Linux/Mac. Macs ship with the bash shell by default. Before you run out and buy a book on Bash, remember that Google is your friend and that you can quickly learn Bash using free resources on the web.
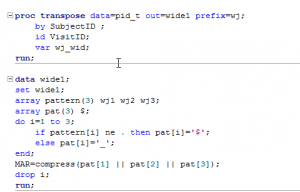
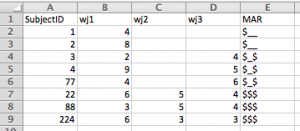
Using bash, you can turn something that would require the manipulation of a lot of files into something quick and easy. For instance, lets say that you need to use FSL’s BET (Brain Extraction Tool) to skull strip a hundred brains. In this case the files follow the pattern – subj001.nii subj002.nii subj003.nii, etc. If you wanted to run bet on each of these, you could do something along the lines of: bet <input file> <output file> <options>. As you can imagine, doing that a hundred times can become very tedious! Scripting to the rescue.
#!/bin/bash
for aSubject in subj*.nii
do
base=`basename $aSubject .nii` #removes the “.nii”, now base contains ex: subj001
bet $base ${base}_brain
done
The script that you can save as a file or just type into the bash shell (Terminal window) and press enter will run through all of the image files you have in the folder that follow our naming pattern and run bet on them.
So scripting is immediately useful, because you can now automate something that would have taken you a long time. But automatic and fast aren’t always the same thing. Insert GNU Parallel. With GNU Parallel, you can run all of these tasks simultaneously (assuming that you have more than one processor in your computer). To do this, we no longer need the for loop and our command becomes this:
ls subj*.nii | parallel bet {} {.}_brain
This command says get a listing of all files in the directory following our naming scheme. Now pipe that listing into parallel. Parallel will then call bet on all of those files piped into it and output them without the extension {.} and add on the suffix _brain. In terms of efficiency, running the shell script above with the for loop took approximately 50 seconds on 9 files. Running the process in parallel took 7 seconds with the same 9 files. That’s it. No hard setup of a cluster, no learning a special programming language. Just parallel.