I’ve written about ROI creation in AFNI a few times before (one, two, three). When we draw ROIs or use 3dUndump, we usually end up with multiple ROIs in one file, each with a different label. In contrast, when we use whereami to create atlas regions, we usually end up with one ROI per file. We can combine those ROIs using 3dcalc or 3dmask_tool. But if we end up with lots of ROIs (more than 7), and we want to combine them into a single file for viewing, this begins to get more difficult. Let me use a simple case, where we wanted to look at the connection between three components of the IFG (Opercularis, Triangularis, Orbitalis) and the fiber tracts that connect these regions to the precentral cortex — because we’re hoping that it’s all about articulation and motor mapping for this particular sample of individuals.
The basics are fairly straightforward. First use whereami to create the ROIs for each of these four regions:
whereami -mask_atlas_region CA_N27_ML:11 -prefix L_IFG_Op whereami -mask_atlas_region CA_N27_ML:13 -prefix L_IFG_Tri whereami -mask_atlas_region CA_N27_ML:15 -prefix L_IFG_Orb whereami -mask_atlas_region CA_N27_ML:1 -prefix L_Precentral
Notice that I used the numeric indicator to get the regions from whereami, but I could have also typed out the region names. Next I want to combine these regions into a single ROI while giving them a unique number for later use with 3dROIstats or 3dTrackID. From previous posts, you know that I could use 3dcalc to do this:
3dcalc -byte \ -a L_IFG_Op+tlrc \ -b L_IFG_Tri+tlrc \ -c L_IFG_Orb+tlrc \ -d L_Precentral+tlrc \ -prefix allROIs_byte \ -expr 'a+2*b+3*c+4*d'
Notice that I used -byte to force the output to be of datum byte, which is required for 3dfractionize. You can see that this 3dcalc syntax gets arduous if you have a large number of ROIs. The alternative of the day is to use 3dTcat and 3dTstat to combine the ROIs:
3dcalc -a L_Precentral+tlrc -prefix zeroDataset -expr'0' 3dTcat -prefix tmpROIs zeroDataset+tlrc L_*.HEAD 3dTstat -argmax -prefix allROIs tmpROIs+tlrc.HEAD rm tmpROIs+tlrc.* 3dcalc -byte -a allROIs+tlrc -expr 'a' -prefix allROIs_byte
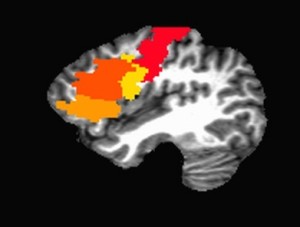
First we create a dataset that just has zeros, then we use 3dTcat to concatenate the zero dataset and all of the ROIs into a single dataset (similar to 3dbucket). And then we use 3dTstat to assign numbers to the ROIs based on where they appear in the sequence of the concatenated files. Finally, I use 3dcalc to turn the dataset into a datum of byte again. We can then view the ROIs in AFNI:
Hopefully this trick saves you substantial amounts of time when you have lots of ROIs in your particular dataset.