In the past I’ve shown how to use 3dttest++ to do one-sample, paired, and two-sample t-tests for whole-brain maps in AFNI. Occasionally there is an instance where you want to quickly generate a series of t-tests for all participants at once. Most people (including me until recently) would simply loop over their gen_group_command.py script several times (with different sub-brick inputs each time) to accomplish this. Another option is to use both the “short form” and the -brickwise option for 3dttest++. The great thing about the short-form is that you can use wildcards. So if you have every datafile in the same directory, you can do something like this:
3dttest++ -prefix all_t -brickwise -setA Subject??.stats+tlrc.HEAD
Which will conduct one t-test for every sub-brick in your input datasets. Which of course means that you need to 1) have all of your input datasets the same size (# sub-bricks) and 2) only include conditions of interest. Both of these requirements usually mean using 3dbucket to extract only the coefficient values as so:
3dbucket -prefix Subject01.stats+tlrc FullSubject01+tlrc.HEAD[Cond1#0_Coef, Cond2#0_Coef, Cond3#0_Coef, Cond4#0_Coef]
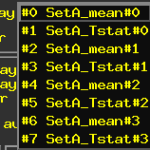
The output of the 3dttest++ with -brickwise on the datasets prepared by 3dbucket will look like the following. You’ll notice unlabeled outputs, and one for each input sub-brick.
At this point you might want to rename your sub-bricks to easily remember what they were later, to name just the first two:
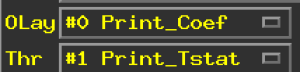
3drefit -sublabel 0 'Print_Coef' all_t+tlrc. 3drefit -sublabel 1 'Print_Tstat' all_t+tlrc.
Which results in the following newly relabeled sub-bricks.
And there you have it! A quicker way to analyze your data without the need to loop over a script repeatedly. Though I have to say that you should be careful because mistakes can happen and you might label something incorrectly and skew your interpretation of some results!