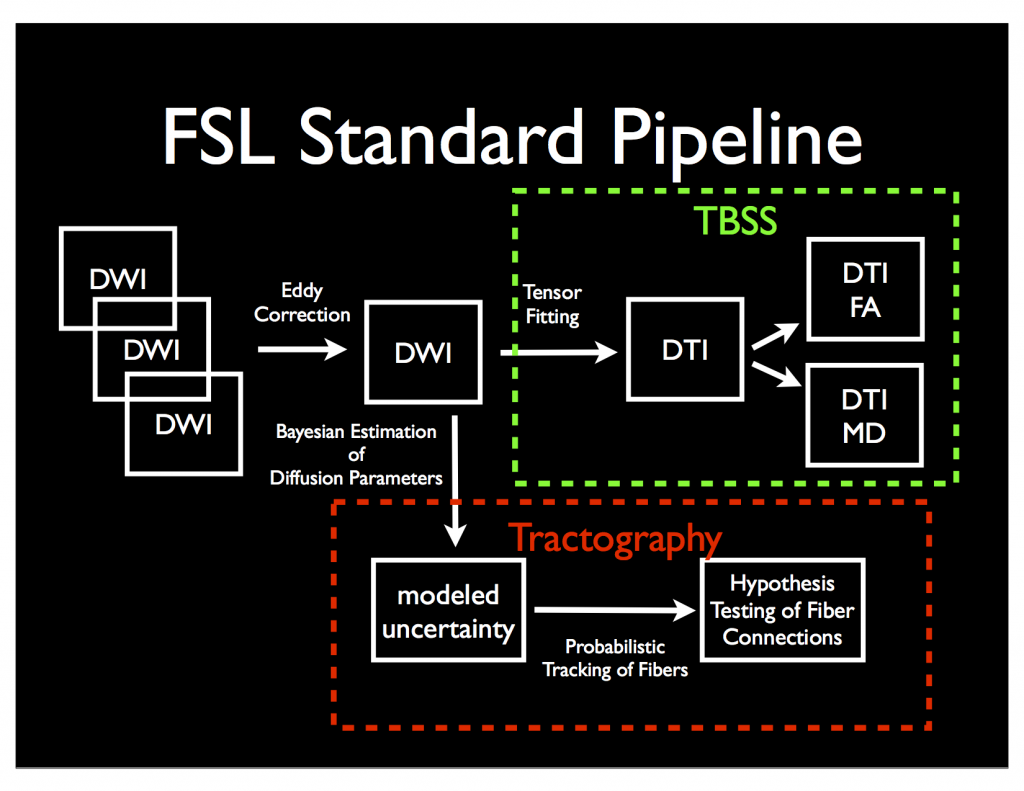
FSL has a complete pipeline for converting Diffusion Weighted Images (DWI) to Diffusion Tensor Images (DTI) using one of two pipelines: 1) “Tract-Based Spatial Statistics” or 2) Probabilistic Tractography (via Bedpostx and probtrackx). The graphic below shows a rough overview of each.
The analysis with FSL is fairly straight forward. If you wish, you can run the FSL Diffusion Toolkit via the Graphical User Interface (GUI) by typing FSL and then clicking FDT or by launching FDT directly – type Fdt_gui into the terminal on a mac, or omit the “_gui” on linux. Alternatively you can process the data via the command line, which ends up being easier to batch across multiple subjects! I’ll print the command lines below along with a step by step guide. We’ll start with the TBSS pipeline and I’ll save the bedpostx and probtrackx for another post.
Step 0: Organize all of your subjects into consistent naming schemes and into separate folders. If you used dcm2nii to convert your DICOM files to NIFTI format, it will conveniently also generate a bvec and bval file for each DWI NIFTI file.
Step 1: Eddy Correction
eddy_correct subject1.nii.gz subject1_ec.nii.gz 0
Step 2: Create Mask – necessary for tensor fitting!
bet subject1_ec.nii.gz subject1_ec_mask.nii.gz -m -n -R
Step 3: Fit Tensors
dtifit -k subject1_ec.nii.gz -o subject1_tensor.nii.gz -m subject1_ec_mask.nii.gz -r subject1.bvec -b subject1.bval -V
Step 4: TBSS Preprocessing
More detailed instructions available from FSL Wiki!
First, copy all of your FA files into one folder. Next, take a look at your data files, I recommend naming them in a way that the groups stick together. Perhaps adding a prefix of “Control_” to one group and “Exp_” to the other. Use design_ttest2 to layout your design.mat and design.con files.
Next, run tbss_1_preproc *.nii.gz inside that folder. Followed by tbss_2_reg to register all images to the template. tbss_3_postreg will actually apply the nonlinear registration. tbss_4_prestats will threshold the images. Finally you can use randomise to run your analyses.