It’s been a while since I’ve touched on this topic (Part 1, Part 2). Recently the topic of doing t-tests has come up a lot in the area around where my desk is situated. In vastly over simplified thinking, I think the three most common approaches to using t-tests in MRI data are as follows:
- One-sample t-test – identify significant activation exists compared to zero.
- Two-sample t-test – identify where two groups differ in activation
- Paired-sample t-test – identify where two conditions differ
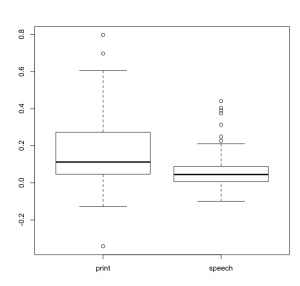
It’s important to note that these group level t-tests can be performed on single activation coefficients or on contrasts at the single subject level. Be careful, this can get complicated very quickly if you’re asking for how two groups differ on a difference between two conditions. Avoiding for a moment the hot debate about correct statistics to use, let’s just focus on using 3dttest++ via gen_group_command.py to perform these three analyses. For anyone not yet familiar with gen_group_command.py, it is a python script that easily generates syntax to run 3dttest++, 3dMEMA, 3dANOVA2, and 3dANOVA3. It’s well worth learning! That said, for the examples in this post, I have a dataset where participants either read print in the scanner or listened to speech.
In order to get the subs_betas used below, I ran 3dinfo -verb subject01+tlrc. and found that I had Coef (stands for coefficients) for print and speech with the format ‘print_GLT#0_Coef’ ‘speech_GLT#0_Coef’, I could also use the numbers corresponding to those sub-briks, but I think using names is safer and would encourage you to do the same! I’ve also included the -set_labels command, which will label the output sub-briks in useful ways for interpreting the analyses later on!
One-sample t-test
If I wanted to see what areas of the brain activate in response to one of these conditions, I would choose to do a one-sample t-test. For example using just print, the gen_group_command would look something like this:
gen_group_command.py -command 3dttest++ \ -write_script ttest_print.tcsh \ -prefix print_results \ -dsets subject??.stats+tlrc.HEAD \ -subs_betas print_GLT#0_Coef \ -set_labels print
If I wanted to look at the difference between print and speech, I could also use a one-sample t-test, if I thought ahead and included that GLT in the single subject analysis. It would look something like this:
gen_group_command.py -command 3dttest++ \ -write_script ttest_print-speech.tcsh \ -prefix print-speech_results \ -dsets subject??.stats+tlrc.HEAD \ -subs_betas print-speech_GLT#0_Coef \ -set_labels print-speech
Paired-sample t-test
If I didn’t think ahead and put the print-speech contrast in the single-subject analysis, I could instead use a paired-sample t-test (because they are the same subjects). Notice the two separate Coef’s in the subs_betas line and the two uses of the -dsets option!
gen_group_command.py -command 3dttest++ \ -write_script ttest_print-speech.tcsh \ -prefix print-speech_results \ -dsets subject??.stats+tlrc.HEAD \ -dsets subject??.stats+tlrc.HEAD \ -subs_betas print_GLT#0_Coef speech_GLT#0_Coef \ -set_labels print speech \ -options -paired
Two-sample t-test
Now if I had two groups of subjects and I wanted to look at how those two groups differed on brain responses to print, I would use a two-sample t-test.
gen_group_command.py -command 3dttest++ \ -write_script ttest_print-speech.tcsh \ -prefix print-speech_results \ -dsets Group1/subject??.stats+tlrc.HEAD \ -dsets Group2/subject??.stats+tlrc.HEAD \ -subs_betas print_GLT#0_Coef \ -set_labels print
Don’t forget to run the scripts!
It’s important to note that running gen_group_command.py just sets up the script for you. Here my individual subject files are labeled subject01, subject02, etc. You can look at the generated tcsh script and make sure that the command looks something like what you wanted. Then you can execute the script by typing: tcsh MyScript.tcsh, where “MyScript.tcsh” is one of your t-tests that you created with gen_group_command.py.
Don’t forget you can mask too!
Masking allows us to hide activation outside of where we might want it. But while it makes pretty pictures, it can also cause you to miss problems in your data. When I first run group analyses, I usually don’t mask the data, this allows me to see if there are glaring problems (like huge activation outside of the brain). With any of the snippets above, you can pass gen_group_command.py the flag -options and after that a -mask and the path to your mask. You could easily run the gen_group_command.py twice, once with and once without the mask. As an alternative, you could mask the data after the fact using 3dcalc.
3dcalc -a print-speech_results+tlrc -b MyMask+tlrc -expr 'a*b' -prefix masked_print-speech_results
I always put the dataset first and the mask second, because 3dcalc will by default output a dataset in the same datum (float, byte, short) of the first dataset passed to it. Try switching them and you’ll be greeted by a warning and data that doesn’t look the way you think it should!