After my first year of blogging, I ran a “One Year Review” piece that covered all of the articles that I wrote in that first year. And that article tends to get hit by search engines fairly often since it was published on December 3, 2013. Sure, I meant to go back and do a “Two Year Review” piece in 2014, but I clearly failed to do that! It wasn’t until yesterday when a kind reader emailed in to ask why either 1) everything wasn’t converted over to the wiki yet, or 2) there weren’t more review pieces that had helpful links to major articles on one topic. The answer to both of these is that things have been a bit crazy busy here, and I’ll try to do better! Nevertheless, since the reader specifically asked for DTI articles, I’m going to do a topic review here calling out all of the articles that cover some semblance of DTI data analysis and perhaps that will give you inspiration for reading some of them! I’ll also try to wrap up with a “where are these posts going in the future bit,” unless I get distracted by a puppy sometime between the start of this article and midnight.
As with all things in imaging, you have a large number of possible avenues of analysis. The first question most people ask is “what software should I use!?!”. The answer is usually “it depends!”. I’m not trying to dodge the question, it really does depend, to some folks it may depend on the type of analysis that you wish to do. If you’re interested in doing “whole-brain” style analyses on DTI data, you might be interested in FSL‘s TBSS package or Freesurfer’s Diffusion Analysis tools. If you’re interested in doing tractography, you might turn to AFNI’s FATCAT (PDF) implementation or FSL’s bedpostx/probtrackx tools or Freesurfer’s TRACULA pipeline! And these are only a quick partial list of the tools you could choose from for each analysis type! You could read this article for a partial overview of tools. Or, after clicking all of the links above, you might checkout Dipy, TrackVis’ Diffusion Toolkit, or Camino! If you’re interested in packages that take care of a lot of preprocessing and artifact detection/correction, you might look more at TORTOISE or DTIprep. Or you might do the analyses in a tool that you’re already familiar with to lessen the learning curve or perhaps to more easily integrate your DTI data with your functional and structural data analyses! Whatever your choice may be, there are tutorials for some of these below!
AFNI
Part 1 – Registering DWI data to anatomical & Fitting Tensors
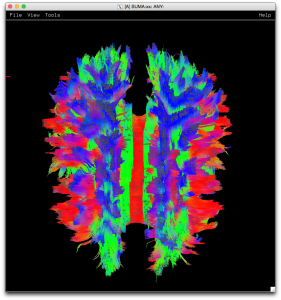
Part 2 – Basics of Deterministic Tractography & Visualization in SUMA
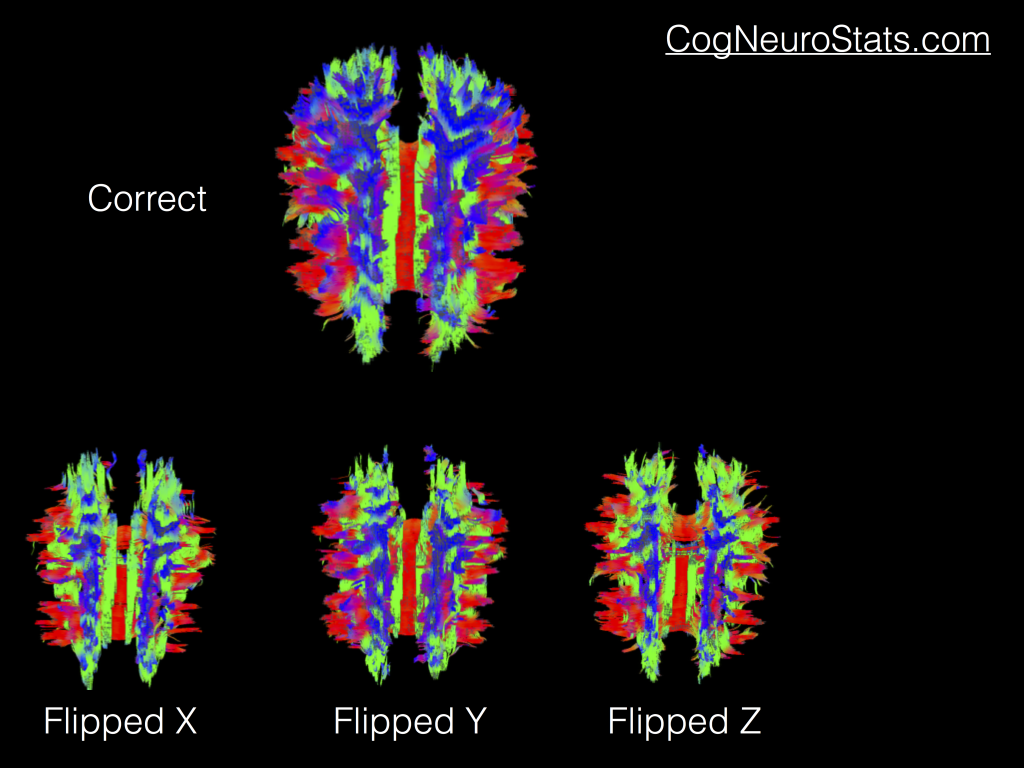
Part 3 – More visualizations and the importance of flipping B-Vectors
Rotating b-vectors – A script using AFNI tools to rotate B-Vectors due to movement parameters
FSL
Part 1 – Basics of DTI in FSL for tensor fitting and Tract-based Spatial Statistics (TBSS)
Part 2 – Visualization of DTI results in FSL’s viewer (aptly named FSLView)
TORTOISE
Part 1 – Preprocessing Diffusion data using TORTOISE’s DIFF_PREP tool
Part 2 – Fitting tensors using TORTOISE’s DIFF_CALC tool
Part 3 – Combining blip-up blip-down diffusion data in TORTOISE for optimal results
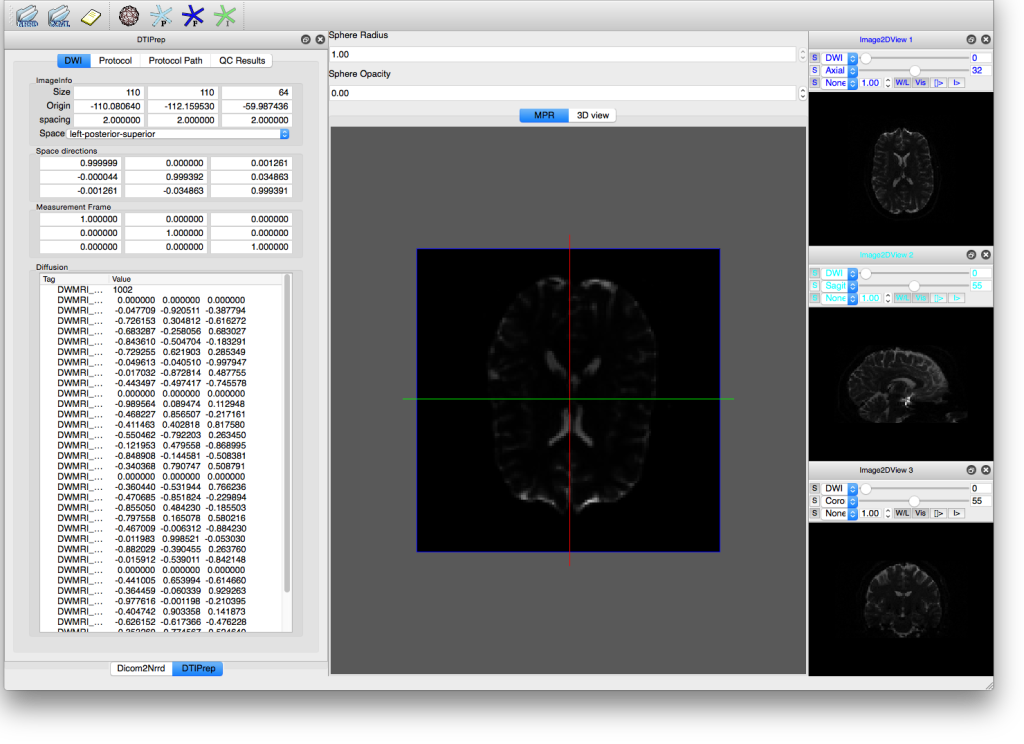
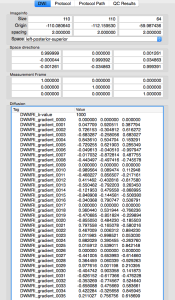
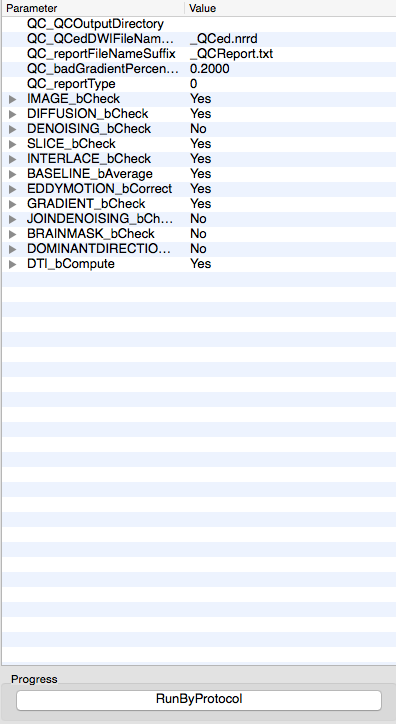
DTIprep
Part 1 – HowTo preprocess diffusion data using DTIprep
Part 2 – An example script to automate using DTIprep
Happy processing!